Research
Genome organization in the nucleus


The organization of the genome in the eukaryotic nucleus is crucial for a multitude of functions, ranging from transcription regulation to the maintenance of chromosome order to ensure proper mitosis [Vivante 2018].
The organization is achieved through a hierarchical set of structures and mechanisms; it includes nucleosomes that shorten the chromosomes, topologically associated domains that assists in gene regulation as well as chromosome territories that ensure that the molecular organization is maintained.
We lately analyzed the elastic properties of the chromatin [Vivante 2020], and the lamin A distribution along the cell cycle [Vivante 2021].
We explore the nucleus organization by using a multitude of methods, including the following:
-
Fluorescence correlation spectroscopy (FCS)
-
Continuous photobleaching (CP)
-
Fluorescence recovery after photobleaching (FRAP)
-
Life-time imaging (FLIM)
-
Super-resolution microscopy
-
Force dynamics response in live cells
DNA origami for biophysical studies
DNA origami was developed during the last decade for numerous applications. We have developed various DNA origami structures for studying the physical properties of DNA and for biosensing.
We used special designs that includes single-stranded DNA and we measured the persistence length of DNA, [Roth 2022] [Roth 2019].
This allowed us to achieve an accurate determination of the end-to-end distance of each ssDNA segment, and by fitting the measured distribution to the ideal chain polymer model, we measured an effective persistence length of 1.98 ± 0.72 nm.
We also use DNA origami structures for developing methods for multiple viral detection.
See late publication [Roth 2022].

Proteins and DNA dynamics

Biophysical processes primarily involve diffusion or random motion of some kind. For example, when a protein moves in the cytoplasm, it normally diffuses out. In addition, molecules seldom moves in some kind of a potential well, and they react with other molecules. These three elements - diffusion, reaction, and motion in a potential, determine the dynamics of the molecules; therefore, they are important for a numerous biological processes, including metabolism, the pace of growth, signaling, and more.
A good example is that of a particle trapped in optical or magnetic tweezers, or a particle tethered to DNA (see the figure). We developed a method for studying such a system based on tethered particle motion (TPM) as explained in the next section. We also developed a formalism for analyzing the dynamics based on Smoluchowski’s equation in vitro or on Langevin formalism for describing the dynamics of chromatin in the nucleus.
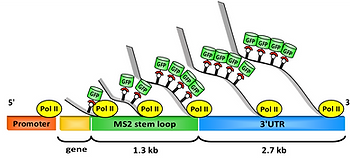
Molecular motors are also of crucial importance, and play an important role in nature, such as the motion of RNA polymerase along the DNA during transcription. We studied the pace of this process in live cells (see the figure) and found that the transcription dynamics of a single gene can be described with a constant rate for all transcribing polymerases, whereas cell population transcription rates follow a fat-tailed distribution.
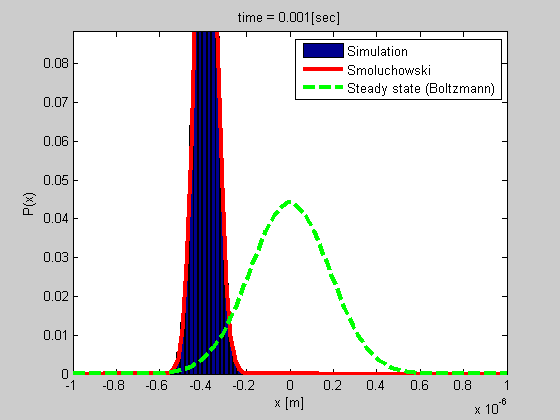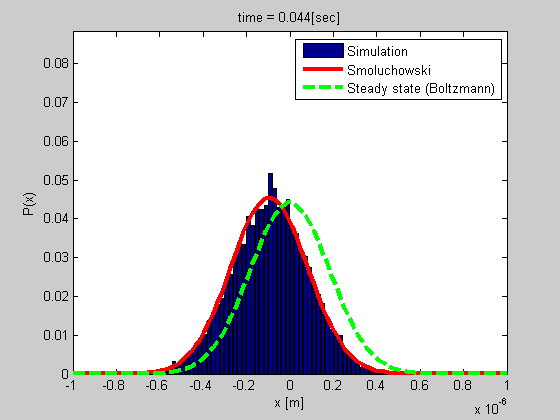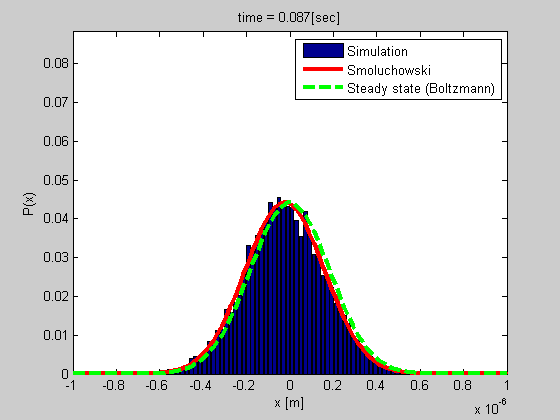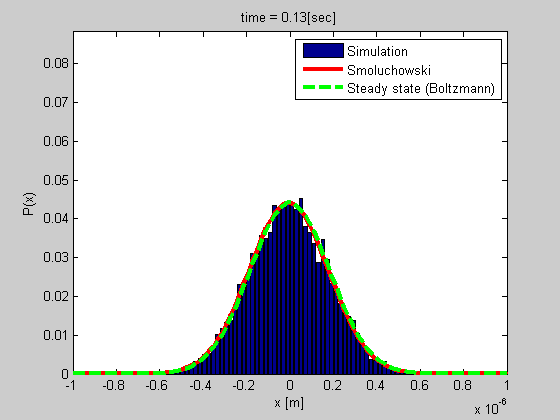
Single detection methods
We use tethered particle motion (TPM), where a nano gold-bead is attached to one end of the DNA, whereas the other end is attached to a glass surface. The DNA molecule diffuses under the restriction applied by the DNA. The bead position is observed through its scattering with a precision that is better than 10 nm. The distribution of the bead positions results from the polymer properties, which can change if proteins are attached to it.
Using TPM, we study the interaction of DNA and proteins, the dynamics of the tethered particle and more. For examle, we found that DNA consists of two different curvature distributions and that the measured persistence length is determined by their interplay. As a result, the effective persistence length adopts a bimodal property that depends primarily on the HU concentration [Nir 2011].
We also used TPM for studying the behavior of short DNA molecules under shear flow conditions, in the proximity of a wall [Nir 2018].


Spectral imaging & cancer detection

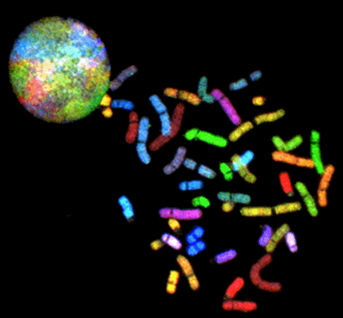
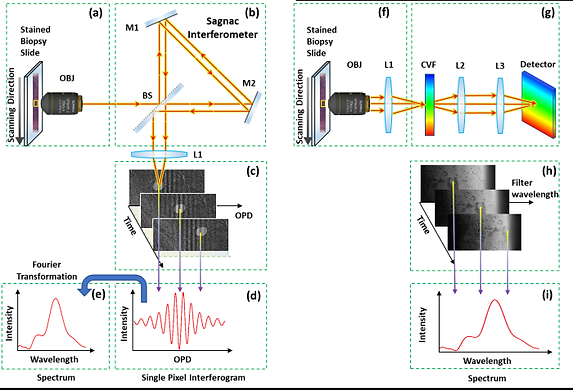
The spectral content of a sample provides important information that is not detected by the human eye. The spectrum contains information about the material being observed. Thus, measuring the spectrum at each point of a sample is important for a large range of applications [Garini 2013]. We developed a novel method for scanning very large spectral images of microscopy samples. This system is based on capturing information while the sample is being scanned continuously ‘on the fly’ [Lindner 2016].
Using spectral imaging, we previously developed a method for screening chromosomal aberrations (see the figure) called spectral karyotyping [Schrock 1996]. The method combines spectral imaging with a biochemical kit that allows one to fluorescently label all 24 chromosomes, each with a different combination of fluorochromes. By measuring the spectrum at each pixel of the image, it is possible to identify the origin of the chromosomal material and identify different chromosomal aberrations.
We have now developed a system for cancer detection.
We lately showed that rapid spectral imaging can be used for cancer detection with unprecedented accuracy. This is done with a unique rapid spectral imaging system that we have developed, see [Brozgol 2022]. Rapid spectral imaging measurement therefore can bridge the gap in the machine-aided diagnostics of whole biopsies, thus improving patient care, and expediting the treatment procedure.
The most important spectral differences are observed in the nucleus and they are related to aneuploidy in tumor cells.